This article is about RF Diffusion, ProteinMPNN, and AF2, key tools in in silico drug discovery. By using these tools, you can design potential protein drugs on your own computer. Once you grasp the content of this article, you’ll be able to experience the process of in silico drug discovery using RF Diffusion, ProteinMPNN, and AF2. Give it a try!
macOS Ventura(13.2.1), python 3.9.7, Jupyter Notebook, PyMOL 2.5.4
What is RF Diffusion?
It is a technique developed at Baker lab, a renowned place for in silico protein design.
RF Diffusion now free and open source
RF Diffusion is one of the methods for discovering new bioactive compounds using machine learning algorithms.
In RF Diffusion, the structural information of known bioactive compounds is used as a dataset, and new compounds are generated from this structural information through machine learning algorithms. These generated compounds are ranked based on their predicted bioactivity, and the most promising compounds can be selected.
In simple terms, RF Diffusion is a method that uses machine learning algorithms to create new compounds from known bioactive compounds, providing efficient and accurate results.
What are ProteinMPNN and AF2?
ProteinMPNN is a deep learning-based protein sequence design method that can design new proteins with high accuracy. ProteinMPNN is trained on a protein databank comprising thousands of high-resolution structures.
Alphafold2 is an artificial intelligence algorithm developed to predict the three-dimensional structure of proteins. This algorithm was developed by DeepMind and announced in 2020. Alphafold2 can predict the three-dimensional structure of a protein from its amino acid sequence with high accuracy.
Alphafold2 is attracting significant attention in the field of protein structure prediction due to its high accuracy. To date, Alphafold2 has predicted the structure of many proteins, showing a high correlation when compared with experimentally determined structures. This plays an extremely important role in deepening our understanding of protein structure and function in the medical and pharmaceutical fields.
Here, we will use a Google Colab page created by Sergeyto perform skeleton generation with RFDiffusion, sequence design with Protein MPNN, and validation with AF2.
RFDiffusionによる骨格生成 →Protein MPNNでの配列設計 > AF2によるvalidationを行います。
To proceed with this article, please download PyMOL.
In silico Drug Discovery Using RF Diffusion, ProteinMPNN, and AF2
Let’s go ahead and try in silico drug discovery using RF Diffusion, ProteinMPNN, and AF2!
First, go to this Google colab page.
This time, we will design a binder for MDMX, a protein model with Protein Data Bank (PDB) number 4N5T.
MDMX is a human protein implicated in cancer development. MDMX can inhibit p53, a crucial cancer suppressor. This can potentially interfere with p53’s normal function and contribute to the formation and progression of cancer cells. On the other hand, loss of MDMX is known to cause embryonic developmental abnormalities.
MDMX has potential as a target for cancer therapy. The development of therapies targeting MDMX is expected to reactivate p53 and suppress cancer cell proliferation. The development of this therapy is anticipated to significantly contribute to cancer treatment.
Please follow the steps below as shown.
- Enter a name in the “name” field. In this case, it’s “MDMXbinder”. Do not include spaces.
- Enter the binding site and length of the protein in “contigs”. Here, we enter “A:30”. We are designing a protein of length 30 that binds to chain A of MDMX.
- Enter the PDB number of MDMX in “pdb”. Here, it’s “4N5T”.
- You can check or uncheck the “Display 3D structure” animation, but it’s interesting, so let’s check it!
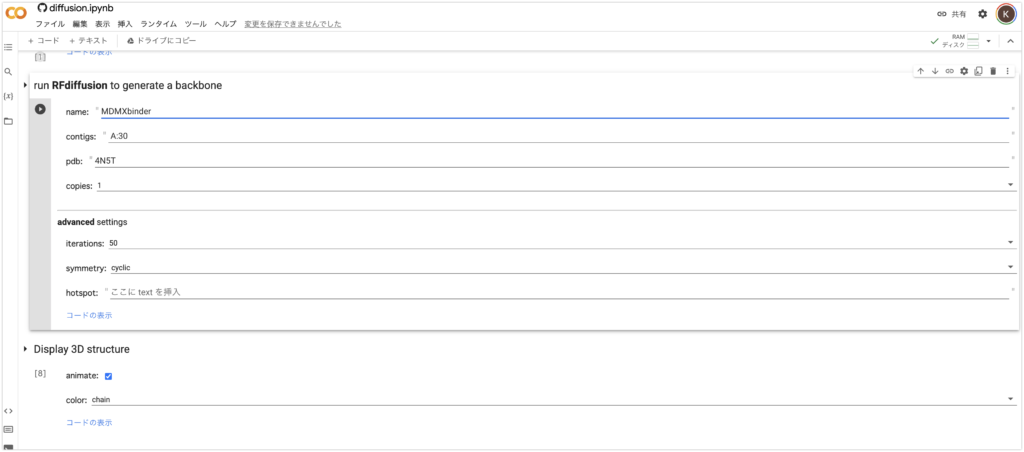
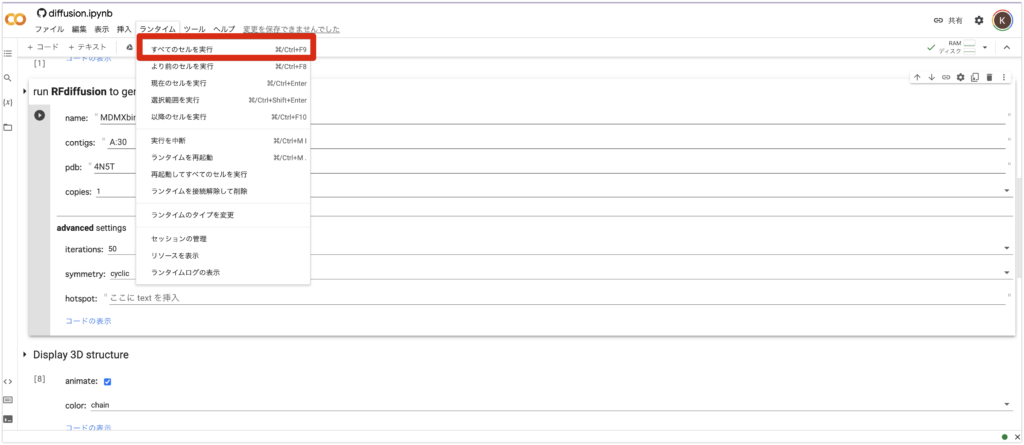
5. Finally, from the “Runtime” tab above, press “Run All Cells”. Then, all you need to do is wait a few minutes.
Generation of Binder Animation
Since we checked the “animate” box earlier, an animation will be generated!
It’s interesting just to look at this, isn’t it?
Result
After a while, the work will be completed and a zip file will be downloaded. Please open the “best.pdb” file in the downloaded files with PyMOL. The most reliable complex will be shown.
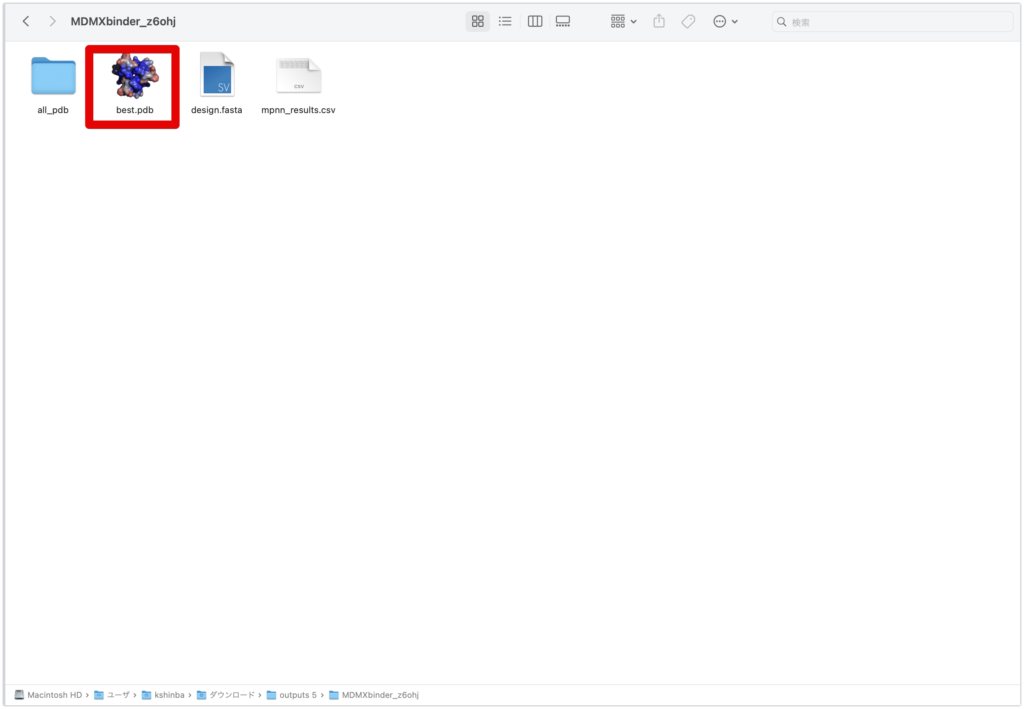
Once open, let’s take a look at the sequence from Display→Sequence.
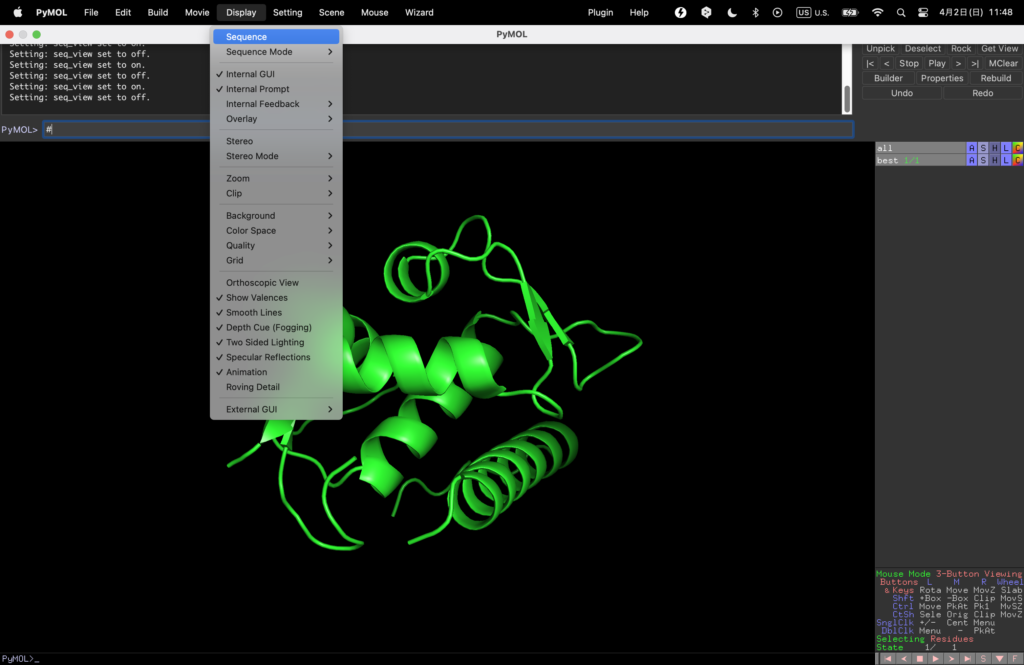
To make it easier to understand, we will color the designed binder red.
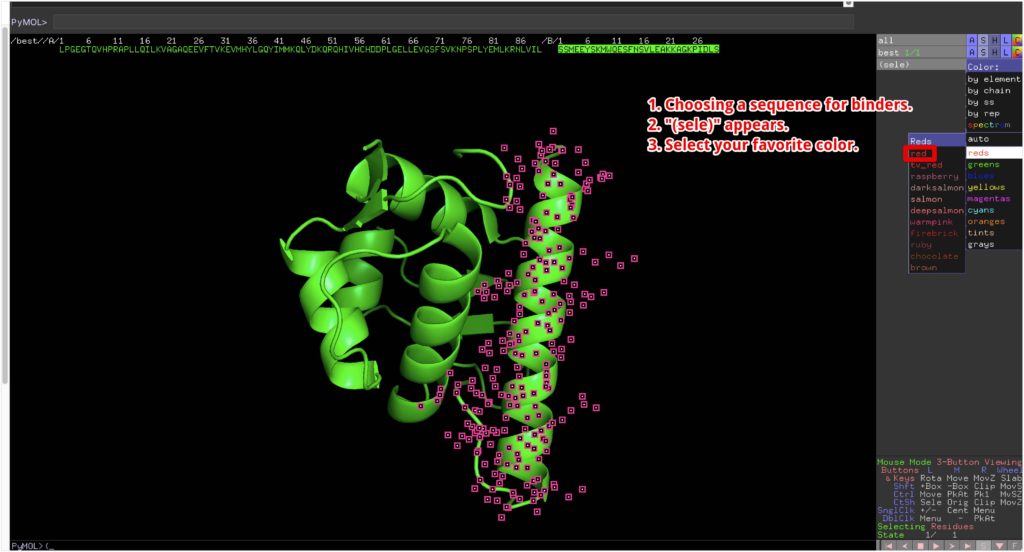
We’ve created a protein with a beautiful α-helix structure! The green one is MDMX, and the red one is the protein we designed this time.
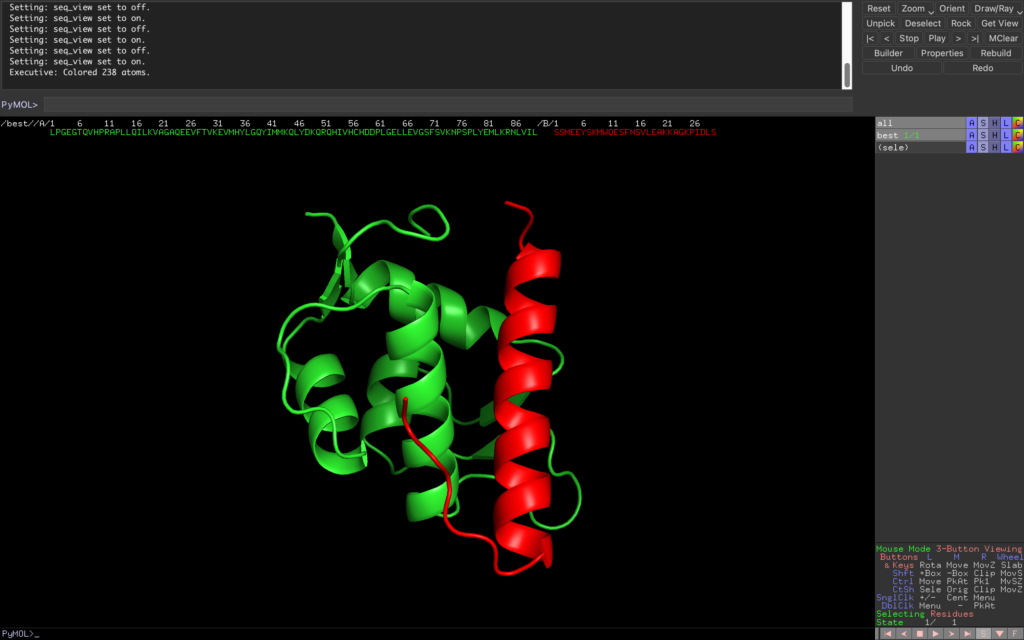
Overlapping with Existing Structures
The MDMX with Protein Data Bank (PDB) number 4N5T is originally in a complex with another binder. Therefore, let’s see how different the binding with the original binder is.
Press File→Get PDB…, enter 4N5T in PDB ID and download the original MDMX and its complex with the binder.
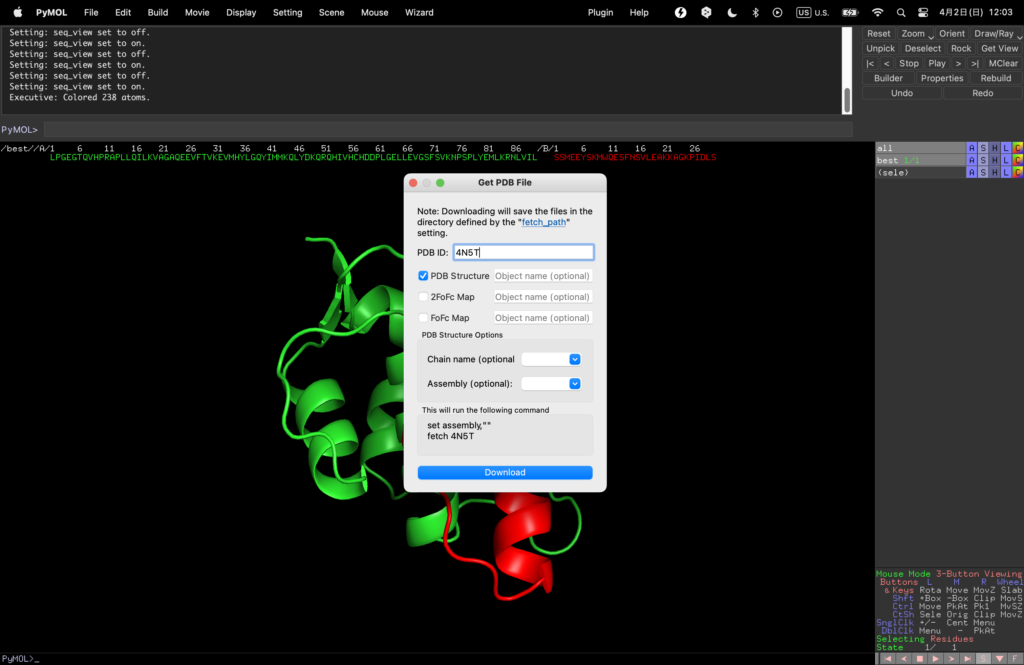
Change the color of the newly emerged MDMX binder (blue here).
Remove water molecules that get in the way by selecting 4N5T→Hide→waters on the right.
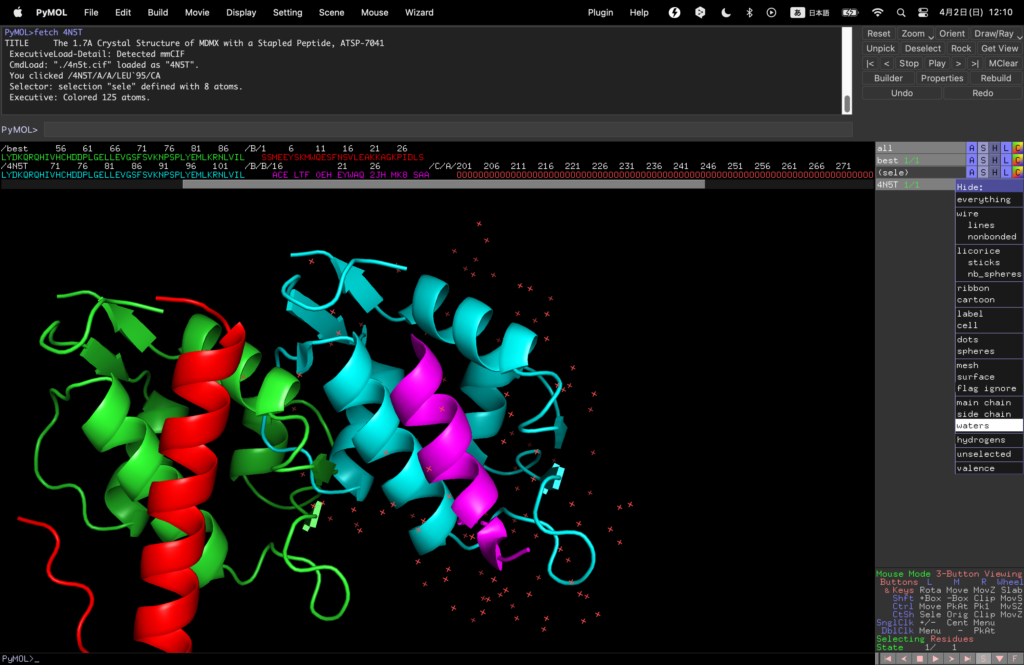
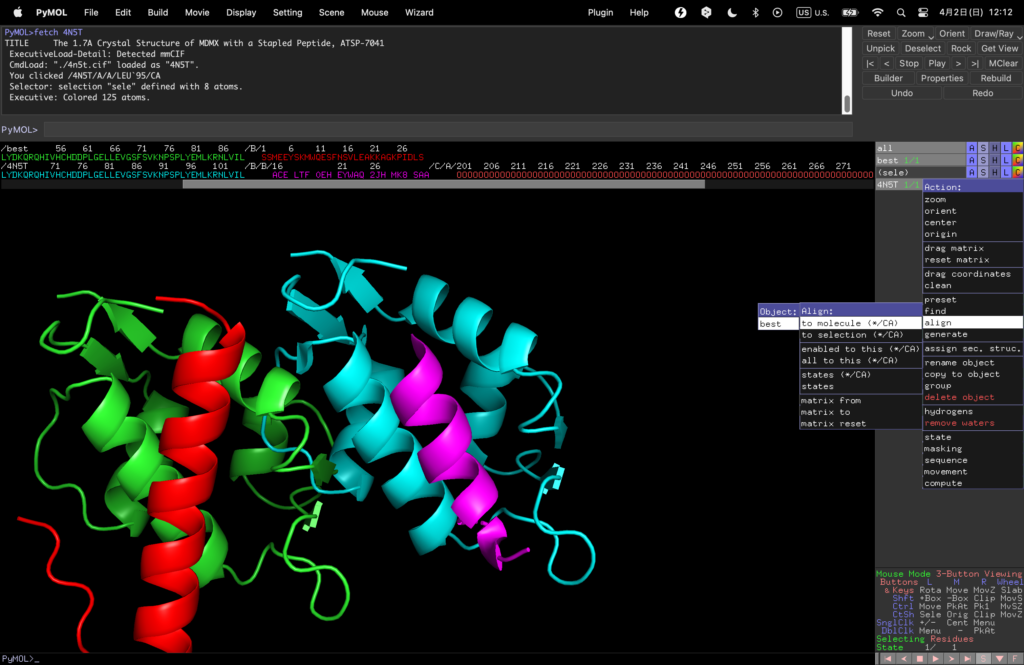
Finally, select 4N5T→Action→align→to molecule→best to align them.
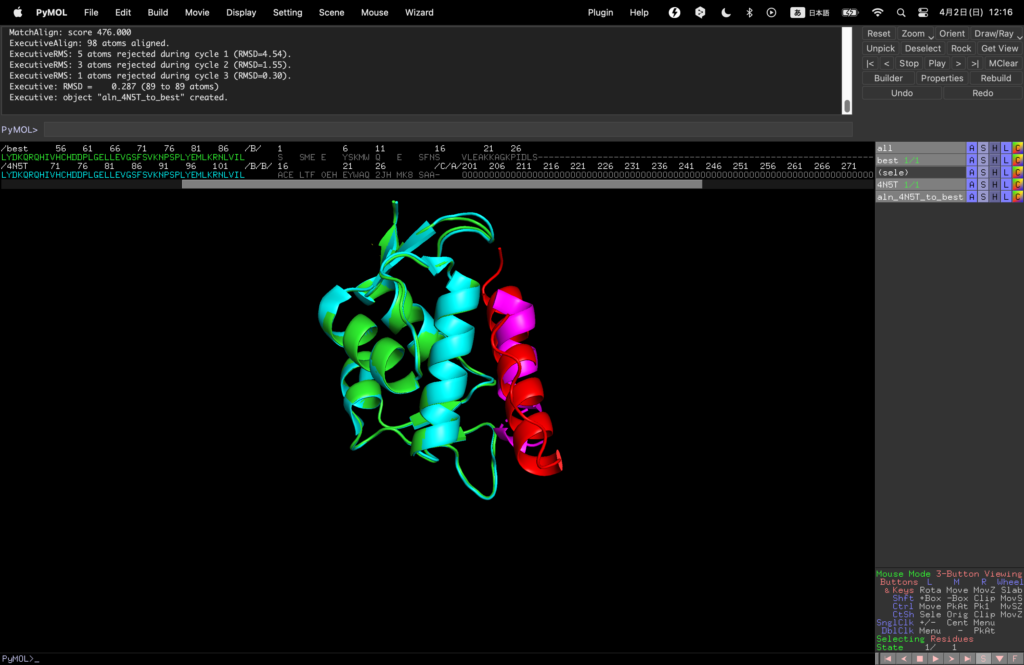
You can see that the binder that binds in the same position as the original binder was designed by this RF diffusion.
Final remark
How did you find it? RF Diffusion, ProteinMPNN, and AF2 allow us to design proteins and engage in in silico drug discovery with remarkable ease. It is indeed a brilliant technology that empowers us to find a binder for any protein of interest. I encourage you all to explore and utilize RF Diffusion, ProteinMPNN, and AF2, and delve into the world of in silico drug discovery. Finally, I would like to express my heartfelt gratitude to everyone involved in developing these amazing technologies.

![[RF Diffusion] Discovery of Protein Drugs using RF Diffusion, ProteinMPNN, and AF2 [In silico Drug Discovery]](https://labo-code.com/wp-content/uploads/2023/07/英語記事用のアイキャッチ画像集.png)